MICROWILD – Deciphering The Microbiome Associated With The Origins Of Agriculture
Coordinators: Dr Pablo García Palacios, Dr Rubén Milla, Dr Manuel Delgado-Baquerizo
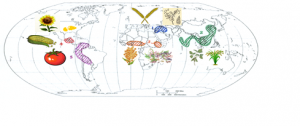
Fig. 1. Map showing the main global centers of domestication and the crop wild progenitors selected across them
We aim to address how domestication has changed the plant microbiome of major crops. This knowledge is needed to optimize the interactions between plants and microbial communities occurring in the rhizosphere, which is crucial to increase global food production. MICROWILD is funded with a grant from Fundación BBVA.
We have secured partnerships with research groups working on wild progenitors of the following crops, and field sampling is taking place during 2019-2020: Wheat (Turkey), Rice (China), Maize (Mexico), Soya (China), Cotton (Mexico), Sunflower (USA), Potato (Bolivia), Tomato (Ecuador), Sorghum (Kenya), Bean (Mexico)
Our main objective is:
1) To identify the core microbiome of crop wild progenitors.
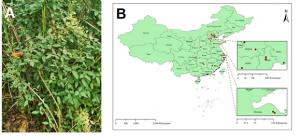
Fig. 2. (A) Population of the wild progenitor of soya (Glycine soja) sampled in China. (B) Map showing the 15 populations of the wild progenitor of soya (Glycine soja) sampled in China
We are coordinating an international consortium to sample the wild progenitors of 10 major crops in their native sites of distribution in the Fertile Crescent, Meso and South America, the Sahel, and South East Asia. Project collaborators are sampling multiple (10-15) populations along a climatic/edaphic gradient for each crop wild progenitor.

